 |

The Reverse Complement tool
Compute the reverse complement of a nucleotide sequence
There are two ways to obtain the reverse complement of a sample with DNA Sequence Assembler:
- Using the 'Reverse Complement' menu
- Using the stand-alone 'Reverse Complement' tool
Both functions support the IUPAC DNA ambiguity code.
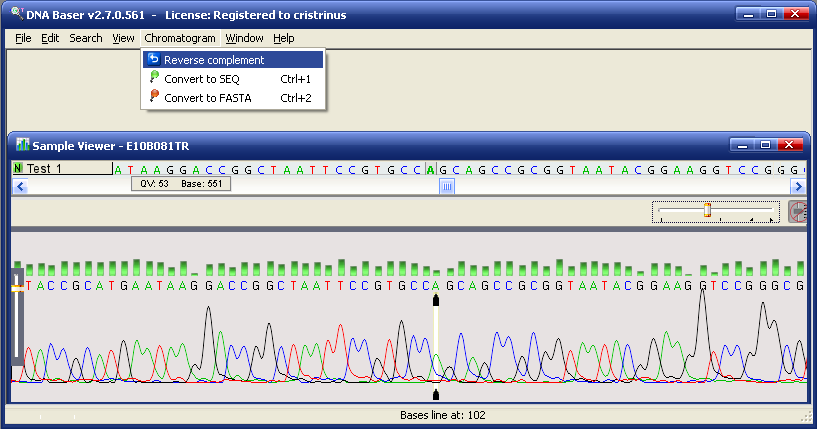
1) The Reverse Complement menu
- Open a sample (Abi, Scf, Fasta, Seq, Txt) in Sample Viewer
- Go
to Sample menu and press Reverse complement
- The RC (reverse complement) of that sample is displayed; in the case of abi or scf sample, the chromatogram is also reversed
2) The stand-alone Reverse Complement tool
- In the main DNA Baser window go to Tasks menu and click the 'Reverse Complement Tool'. The Reverse Complement Tool will open in a new window
- Enter the DNA sequence you want to reverse in the top panel and press the main button
- It's reverse complement will be displayed in the lower panel
Notes
- IUPAC ambiguity codes of the two possible nucleotides are converted as follows: R↔Y, K↔M, W and S are left unchanged.
- Ambiguity codes of the three possible nucleotides are converted as follows: D↔H, B↔V.
- The program preserves upper and lower case. This is useful for marking regions of interest.
- DNA and RNA sequences are converted into reverse-complementing sequence of DNA.
- Unknown/invalid symbols (for example '?') are left unchanged.
- This tool is available for free.

|
 |
