FAQ about confidence scores
(quality values)
DNA Sequence Assembler v3

What is the confidence score (confidence score)?
Definition: The confidence score (also called bases trust information or confidence value or quality value) is a number assigned to each base in chromatogram that shows how much
that base is trusted. A small confidence score means that the predicted base can’t be trusted too much (the base caller may be wrong). The confidence score can
have any value from 0 to 100. A high confidence score
means that the base can be trusted. By default DNA Baser considers a base untrusted if its confidence score is under 25-30.
How important is the confidence score for DNA Sequence Assembler?
DNA Sequence Assembler is the only software on the market that automates the sequence assembly process reducing the required time
with about 1000%.
In order to generate high quality/trustable contigs WITHOUT any human intervention, DNA Sequence Assembly relies on the confidence score (confidence score) information assigned to each
base as follows:
1)
Usually bases of low quality are gathered at the ends of your samples. These clusters of low quality bases are called untrusted regions. When performing
sequence assembly or other DNA sequence analysis, the user have to manually cut
out (trim) those bases from samples, else the assembly may be very poor (too many ambiguities) or even wrong. This is a time consuming process!
DNA Sequence Assembler can automatically trim the untrusted regions before it assembles
the sequences.
2) During sequence assembly, if an ambiguity is encountered, DNA Sequence Assembler will use the confidence score information
to automatically correct the ambiguity for you.
If the confidence score information is missing, DNA Baser will not
automatically trim the untrusted regions. In this case the user will have to inspect the contig an check if the suggestions made by DNA Baser Assembler are correct.
UPDATE
Starting with v4, DNA Sequence Assembler does not need the QV info to be already present in the file anymore!
If the QV info is missing it will compute it using its new state-of-the-art base caller.
Most of the information below is therefore obsolete.
Which are the formats that can store confidence score information?
Only SCF and ABI sample file types can store confidence score information. FASTA, SEQ and TXT files cannot store this information.
If I have ABI or SCF files, it means implicitly they contain confidence score?
Not necessarily. If your sequencing machine is set to generate SCF files, those files will probably (99.9%) contain confidence score information. If your sequencing machine is set to generate
ABI chromatogram files, some of the files may not contain confidence score (confidence score) information. However, your technician can fix this easily by setting the
machine to store confidence scores in your ABI chromatogram files.
What to do in case your ABI files do not have confidence score filed included?
Tell your technician or sequencing company to include confidence scores in your chromatogram files (ABI or SCF). All they chack to do is check a checkbox in the machine’s (software) interface. Anyway, DNA Sequence Assembler v4 will automatically compute the confidence scores.
Why DNA Sequence Assembler does not automatically trim the low quality regions in my chromatograms?
Usually you don't have to manually trim the ambiguous bases (the 'N's) from your sequences. DNA Baser will clean the end automatically for you BUT ONLY IF the confidence scores are included
in the file. SCF files always contain confidence score. ABI files only sometimes have this information included. You can instruct your sequencing machine to always generate ABI files with confidence score included
(you just need to check a checkbox in your machine's interface).
How can I find out if my samples contain information about confidence scores (confidence score)?
With DNA Baser, it is very easy to get this information: double click an ABI/SCF file to open it in DNA Baser. If you see green columns above each base in chromatogram, then your file
has the confidence score field included.

Fig 1. ABI file with confidence score included
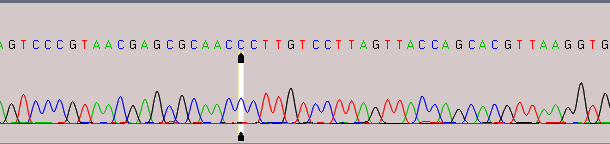
Fig 2. ABI file without confidence score included (green bars indicating the quality of each base are missing)
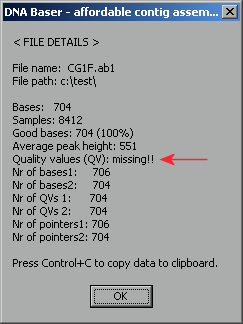
Another way to check the existence of confidence score is to see the properties of a file. Press Control+Enter to see the properties dialog:
If my samples do not contain confidence scores, does it mean that I can’t make a contig?
In most cases, DNA Baser will be able to generate contigs without problems even if your samples does not contain confidence score field. However, in some very rare cases (especially when your samples
are extremely poor), DNA Baser may not succeed in creating contig. In this case, you must manually trim the untrusted ends of your samples/chromatograms.
Can I mix ABI and SCF files?
Sure. You can assemble ABI, SCF, FASTA, SEQ files together.
Can I mix samples containing confidence score with samples that do not contain confidence score (like SCF and FASTA)?
Yes. However, DNA Baser will create the contig in non-confidence score mode. It may be better to totally remove that file from contig than to use it. See the table below:
|
How many files contain confidence score info?
|
Contig will be done in:
|
|
All
|
confidence score mode
|
|
None
|
non-confidence score mode
|
|
Some
|
non-confidence score mode
|
In rare situations, DNA Baser generates poor contigs. Why?
Because your files contain no confidence score information. You may need to trim the untrusted regions manually.
Related topics
|
