|
DNA Chromatogram Explorer
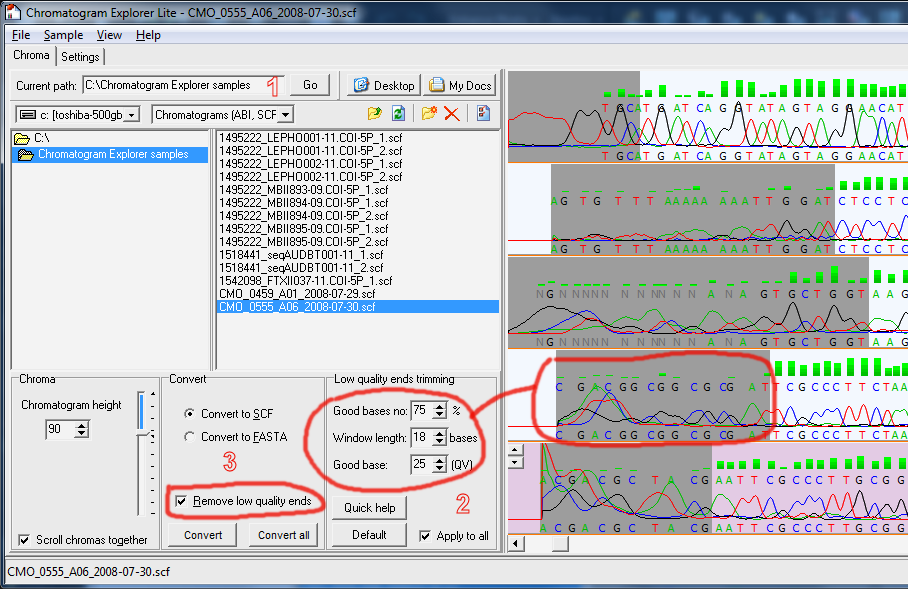
How to automatically trim low quality ends in all chromatograms

With DNA Chromatogram Explorer
you can automatically trim low quality ends to all chromatograms in a folder. To do so we will use the 'Convert' tool.
- Navigate to the folder where your ABI/SCF chromatograms are located
- In 'Low quality end trimming' panel choose how much you want to cut from your chromatograms
- In the 'Convert panel' check the 'Remove low quality ends' and convert all your chromatograms to SCF.
Notes:
- The 'Convert' button converts only the selected chromatogram. 'Convert all' converts all chromatograms in that folder.
- The output will the written in the same folder. Existing files will be overwritten.
- Don't worry if your chromatograms are already in SCF format. Basically in this case the program will only remove the low quality ends and save the file back as SCF.
- The program uses the quality value (QV) data inside the chromatogram to automatically detect low quality regions to cut. If the QV data is missing the program cannot automatically cut the ends. You will have to do it manually in DNA Sequence Assembler. You know that QV data is present when you see a green bar above each base (see image below).
Related topics

|
