 |
Phylogenetic tree
A phylogenetic tree is a tree showing the evolutionary interrelationships between various species or other entities that are believed to have the same common ancestor. A phylogenetic tree
is a form of a cladogram. In a phylogenetic tree, each node with descendants represents the most recent common ancestor of the descendants, and edge lengths correspond to time estimates.
Each node in a phylogenetic tree is called a taxonomic unit. Internal nodes are generally referred to as Hypothetical Taxonomic Units (HTUs) as they cannot be directly observed.
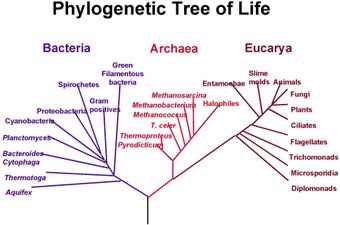
Fig. 1: A speculatively rooted tree for rRNA genes
[Enlarge]
A rooted phylogenetic tree is a directed tree with a unique node corresponding to the (usually inputed) most recent common ancestor of all the entities at the leaves of the tree. Figure 1 depicts a rooted phylogenetic tree, which has been colored according to the three-domain system. The most common method for rooting trees is the use of an uncontroversial outgroup - close enough to allow inference from sequence or trait data, but far enough to be a clear
outgroup.

While unrooted phylogenetic trees can be generated from rooted ones by omitting the root from a rooted tree, a root cannot be inferred on an unrooted tree without either an outgroup or additional assumptions (for instance, about relative rates of divergence). Figure 2 depicts an unrooted phylogenetic tree for myosin, a superfamily of proteins. Links to other pictures are given in the pictures
on the web subsection below.
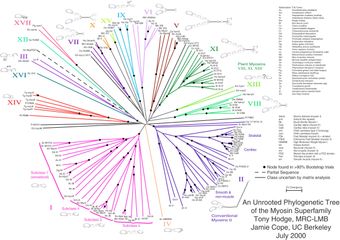
Fig. 2: Unrooted tree of the myosin supergene family
[Enlarge]
The three most used methods to construct phylogenetic trees are: distance-based methods such as neighbour-joining, parsimony-based methods such as maximum parsimony,
and character-based methods such as maximum likelihood or Bayesian inference.
Tree-building methods can be assessed on the basis of several criteria:
- efficiency (how long does it take to compute the answer, how much memory does it need?)
- power (does it make good use of the data, or is information being wasted?)
- consistency (will it converge on the same answer repeatedly, if each time given different data for the same model problem?)
- robustness (does it cope well with violations of the assumptions of the underlying model?)
- falsifiability (does it alert us when it is not good to use, i.e. when assumptions are violated?)
Caveats
- By their very nature, phylogenetic trees cannot represent actual evolutionary patterns and are in fact distorted by, any lateral gene transfer[5] or hybridisation between species
that are not nearest neighbours on the tree before hybridisation takes place. For these reasons, the proposed PhyloCode (see external links below) does not assume a tree structure.
- The phylogenetic tree of a single gene or protein taken from a group of species often differs from similar trees for the same group of species, and therefore great care is needed in
inferring phylogenetic relationships amongst species. This is most true of genetic material that is subject to lateral gene transfer and recombination, where different haplotype blocks
can have different histories.
- When extinct species are included in a tree, they should always be terminal nodes, as it is unlikely that they are direct ancestors of any extant species. Scepticism must apply when
extinct species are included in trees that are wholly or partly based on DNA sequence data, due to evidence that DNA is not preserved intact for longer than 100,000 years.
References
1. Hodge, T. & M. J. T. V. Cope. 2000. A Myosin Family Tree. Journal of Cell Science 113: 3353-3354. See also the Myosin external link below.
2. Woese, C. R. 1998. The Universal Ancestor. Proceedings of the National Academy of Sciences 95: 6854-6859.
3. Maher, B. A. 2002. Uprooting the Tree of Life. The Scientist 16: 18 (Sep. 16, 2002); subscription only
4. Penny, D., Hendy, M. D. & M. A. Steel. 1992. Progress with methods for constructing evolutionary trees. Trends in Ecology and Evolution 7: 73-79.
5. Woese, C. R. 2002. On the evolution of cells. Proceedings of the National Academy of Sciences 99: 8742-8747.
|
 |

